Dayhoff uses shotgun metagenomics—the gold standard in microbiome science—to deliver a complete view of the microbial community in your gut. Our advanced testing detects over 200,000+ bacteria, viruses, fungi, and protozoan parasites, with strain-level accuracy for key species and sensitivity down to 0.005% relative abundance.

At Dayhoff, we use advanced functional profiling to detect which microbial genes are present and how active they are. This helps us understand the real-time activity of your microbiome—what your microbes are actually doing.
Our benchmarks are built on peer-reviewed research and a continuously expanding database of gut microbiome samples. With the world’s largest collection of longitudinal pregnancy and infant microbiome data, Dayhoff is driving the science of early-life microbiome health forward.


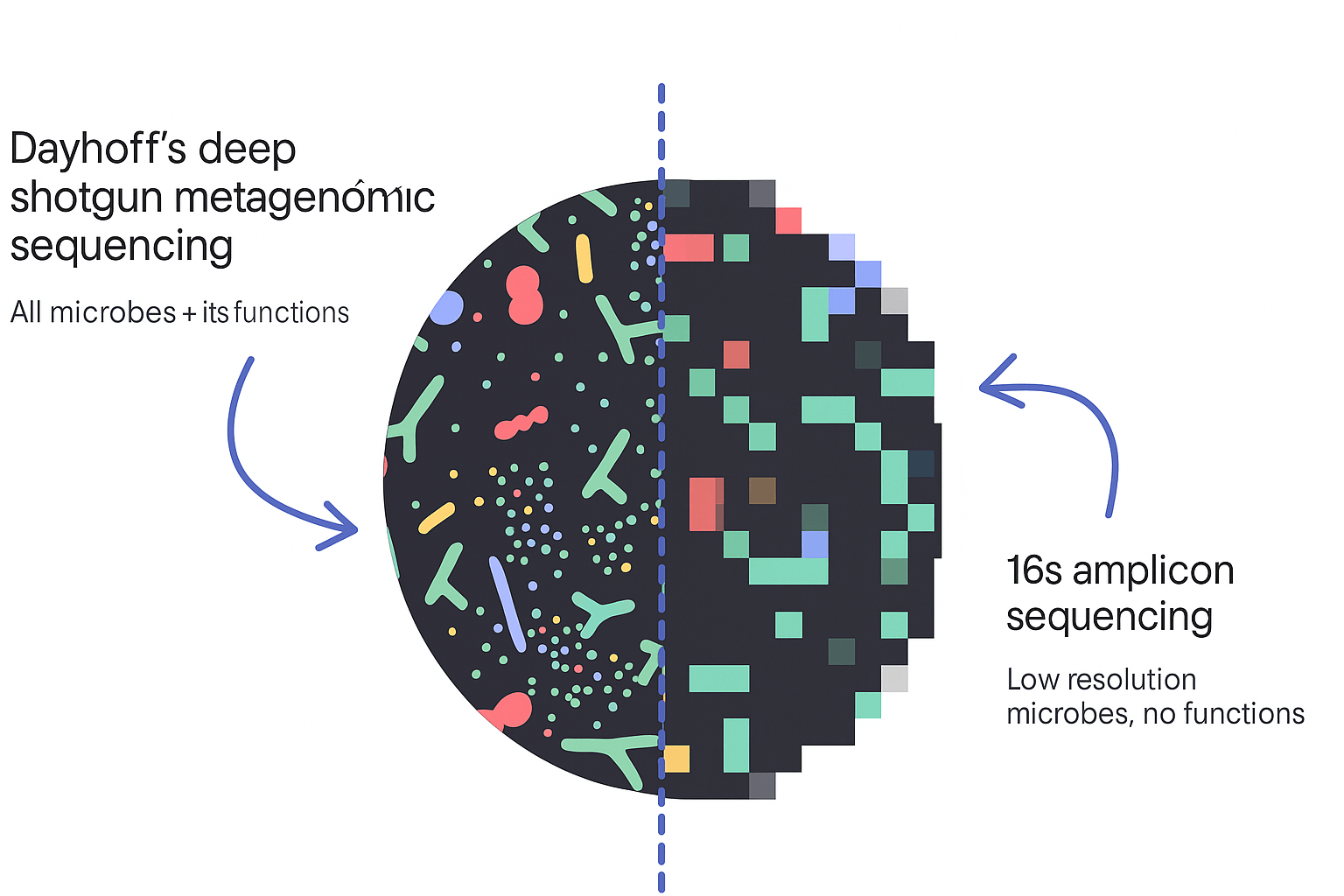
| Feature | Dayhoff | 16s Sequencing |
|---|---|---|
| Results turnaround | 3-4 weeks | 3-4 weeks |
| Price | $599 | $279 |
| Resolution & detail | High – Strain level resolution | Low – Family or genus level |
| Kinds of microbes | Extensive – All bacteria, viruses, fungi, parasites (200,000+) | Limited – Bacteria only (<10,000) |
| Accuracy | High – Limited contamination & false positives | Low – High potential for contamination & false positives |
| % of genome considered | Extensive – Looks at thousands of genes across the entire genome | Low – Looks at only one gene in a section of the genome |
| Future biomarker and therapeutic insights | High | Low |
| Tailored to mom & baby reference ranges | ✅ | ❌ |
| Strain reporting | ✅ | ❌ |
| Antibiotics resistance | ✅ | ❌ |
| Pathogen and toxin recognition | ✅ | ❌ |
| Metabolic function | ✅ | ❌ |
| Microbial capacity | ✅ | ❌ |
| Evidence-based recommendations | ✅ | Basic |
| Require practitioner requisition to order | Not required | Required |
| Expert 1-1 consult call with microbiome expert | ✅ | ❌ |
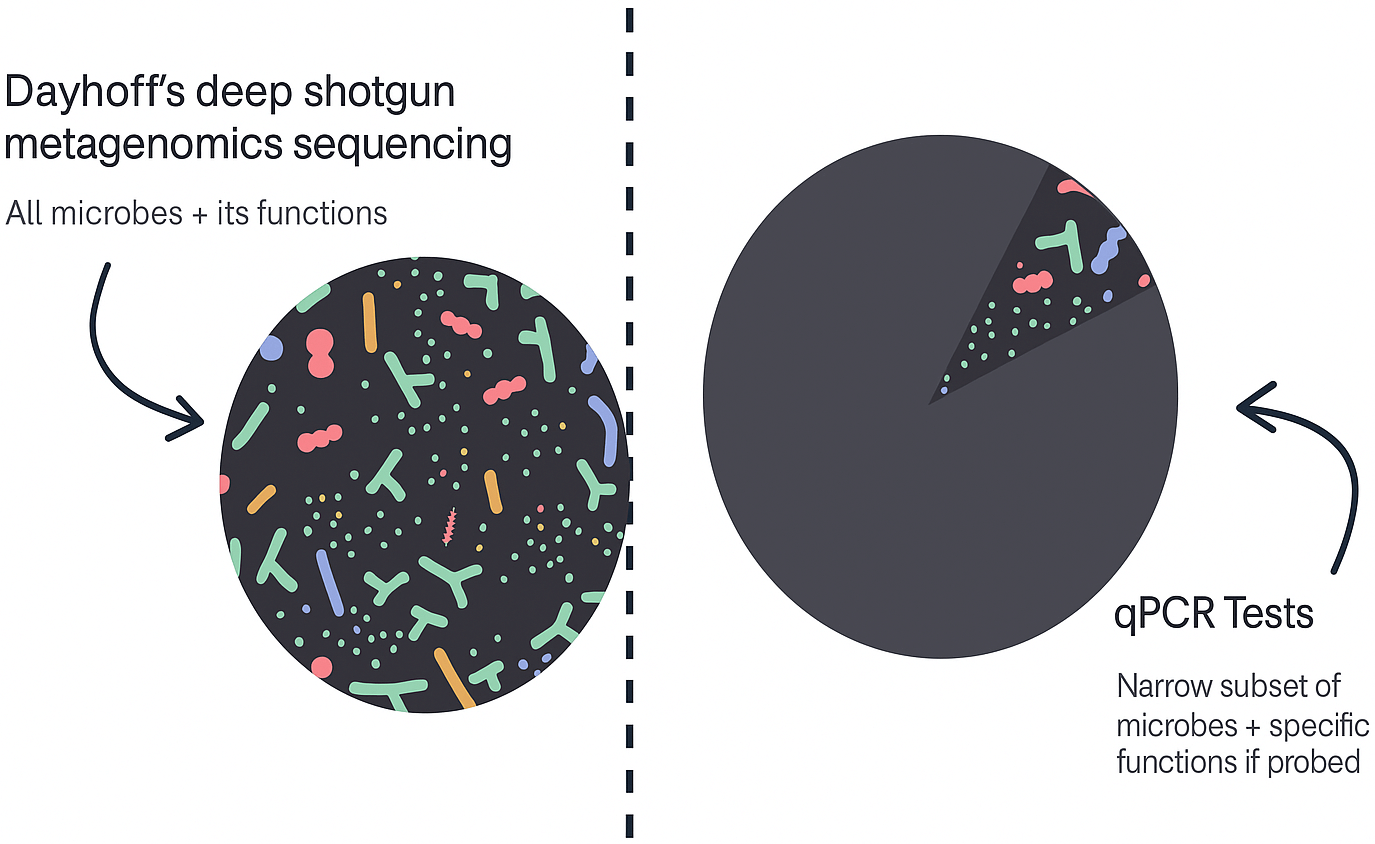
| Feature | Dayhoff | qPCR Tests (GI Map, GI Effects) |
|---|---|---|
| Results turnaround | 3-4 weeks | 3-4 weeks |
| Price | $599 | $400 |
| Resolution & detail | High – Strain level resolution | Limited – Species level, limited report metrics |
| Kinds of microbes | Extensive – All bacteria, viruses, fungi, parasites (200,000+) | Very limited – Probed microbes only (30-80) |
| Accuracy | High – Limited contamination & false positives | High – Highly accurate but not comprehensive |
| % of genome considered | Extensive – Thousands of genes across entire genome | Limited – Multiple genes but not all |
| Future therapeutic insights | High | Low |
| Tailored to mom & baby reference ranges | ✅ | ❌ |
| Strain reporting | ✅ | ❌ |
| Antibiotics resistance | ✅ | ❌ |
| Pathogens recognition | ✅ | ❌ |
| Metabolic function | ✅ | ❌ |
| Microbial capacity | ✅ | ❌ |
| Exact capacity | ✅ | ❌ |
| Evidence-based recommendations | ✅ | ❌ |
| Expert 1-1 consult call with microbiome expert | ✅ | ❌ |
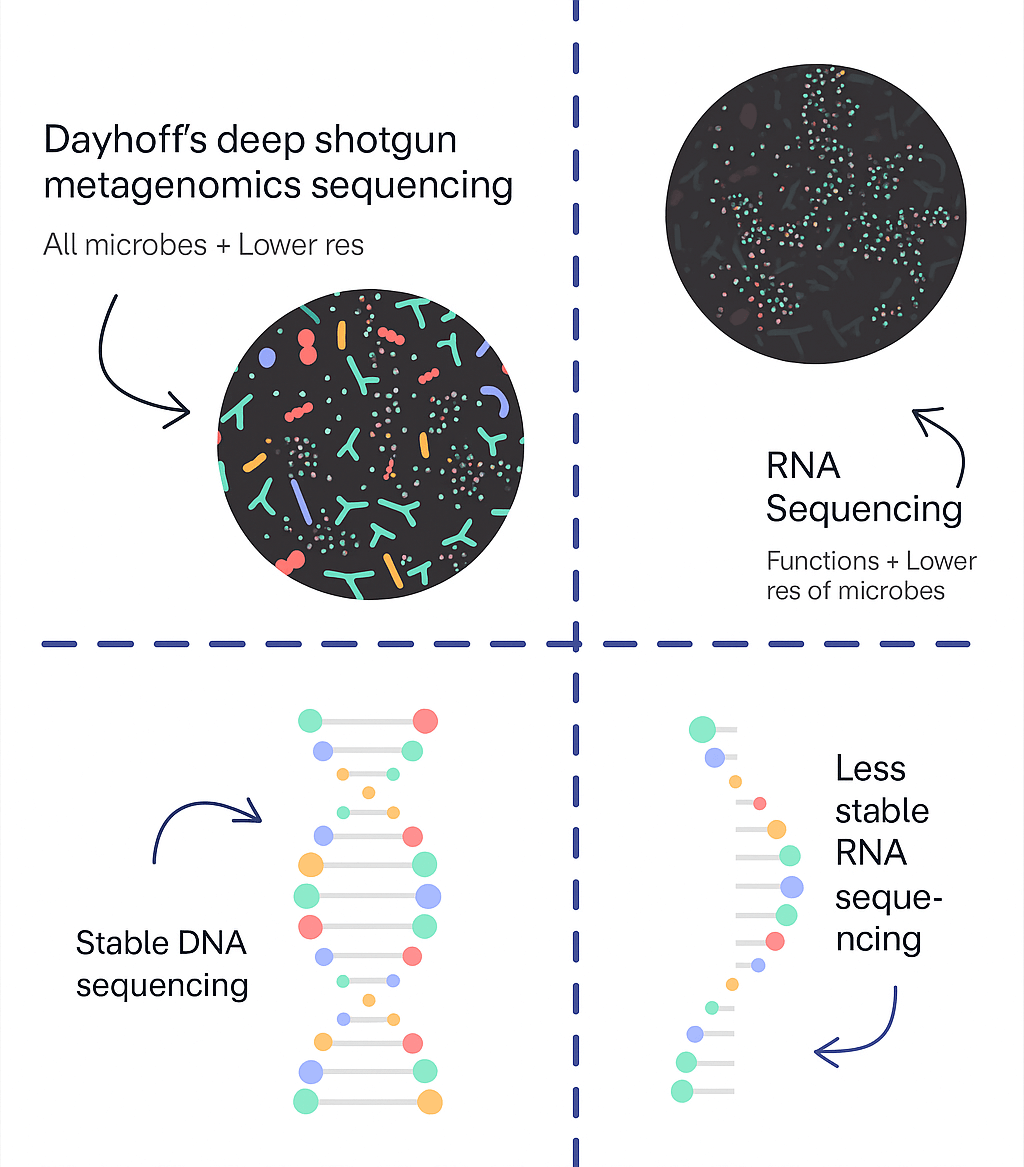
| Feature | Dayhoff | RNA Testing (Viome) |
|---|---|---|
| Results turnaround | 3-4 weeks | 3-4 weeks |
| Price | $599 | $279 |
| Resolution & detail | High – Strain level resolution | Medium – Species level, no reference ranges |
| Kinds of microbes | Extensive – All bacteria, viruses, fungi, parasites (200,000+) | Basic – All bacteria, viruses, fungi |
| Accuracy | High – Limited contamination & false positives | High – Variable based on sample timing |
| % of genome considered | Extensive – Thousands of genes across entire genome | Limited – Uses incomplete genomes |
| Future therapeutic insights | High | Medium |
| Tailored to mom & baby reference ranges | ✅ | ❌ |
| Strain reporting | ✅ | ❌ |
| Antibiotics resistance | ✅ | ❌ |
| Pathogens recognition | ✅ | ❌ |
| Metabolic function | ✅ | ❌ |
| Microbial capacity | ✅ | ❌ |
| Evidence-based recommendations | ✅ | ❌ |
| Expert 1-1 consult call with microbiome expert | ✅ | ❌ |
| Feature | Dayhoff | Metagenomics (Floré, Thorne) |
|---|---|---|
| Results turnaround | 3-4 weeks | 3-4 weeks |
| Price | $599 | $299 |
| Resolution & detail | High – Strain level resolution | Medium – Species level, no reference ranges |
| Kinds of microbes | Extensive – All bacteria, viruses, fungi, parasites (200,000+) | Basic – Bacteria, viruses, fungi, parasites (23,000+) |
| Accuracy | High – Limited contamination & false positives | High – Limited contamination & false positives |
| % of genome considered | Extensive – Thousands of genes across entire genome | Extensive – Thousands of genes across entire genome |
| Future therapeutic insights | High | Medium |
| Tailored to mom & baby reference ranges | ✅ | ❌ |
| Strain reporting | ✅ | ❌ |
| Antibiotics resistance | ✅ | ❌ |
| Pathogens recognition | ✅ | ❌ |
| Metabolic function | ✅ | ❌ |
| Microbial capacity | ✅ | ❌ |
| Evidence-based recommendations | ✅ | Only in-house probiotics |
| Expert 1-1 consult call with microbiome expert | ✅ | ❌ |
| Feature | Dayhoff | Metagenomics (Flore) | RNA Testing (Viome) | qPCR Tests (GI Map, GI Effects) | 16S Test |
|---|---|---|---|---|---|
| Results turnaround | 3-4 weeks | 3-4 weeks | 3-4 weeks | 3-4 weeks | 3-4 weeks |
| Price | $599 | $299 | $279 | $400 | $279 |
| Resolution & detail | High – Strain level | Medium – Species level, no reference ranges | Medium – Species level, no reference ranges | Limited – Species level, limited metrics | Low – Family or genus level |
| Kinds of microbes | Extensive – Bacteria, viruses, fungi, parasites (200,000+) | Basic – Bacteria, viruses, fungi, parasites (23,000+) | Basic – Bacteria, viruses, fungi | Very limited – Limited to probed microbes (30-80) | Limited – Bacteria only (<10,000) |
| Accuracy | High – Limited contamination & false positives | High – Limited contamination & false positives | High – Variable based on timing | High – Accurate but not comprehensive | Low – High potential for false positives & contamination |
| % of genome considered | Extensive – Thousands of genes across entire genome | Extensive – Thousands of genes across entire genome | Limited – Incomplete genomes | Limited – Multiple genes but not all | Low – Looks at only one gene section |
| Future therapeutic insights | High | Medium | Medium | Low | Low |
| Tailored to mom & baby reference ranges | ✅ | ❌ | ❌ | ❌ | ❌ |
| Strain reporting | ✅ | ❌ | ❌ | ❌ | ❌ |
| Antibiotics resistance | ✅ | ❌ | ❌ | ✅ | ❌ |
| Pathogens recognition | ✅ | ❌ | ❌ | ✅ | ❌ |
| Metabolic function | ✅ | ❌ | ❌ | ❌ | ❌ |
| Microbial capacity | ✅ | ✅ | ❌ | ❌ | ❌ |
| Exact capacity | ✅ | ❌ | ❌ | ❌ | ❌ |
| Evidence-based recommendations | ✅ | Only in-house probiotics | ✅ | ❌ | Basic |
| Expert 1-1 consult call with microbiome expert | ✅ | ❌ | ❌ | ❌ | ❌ |
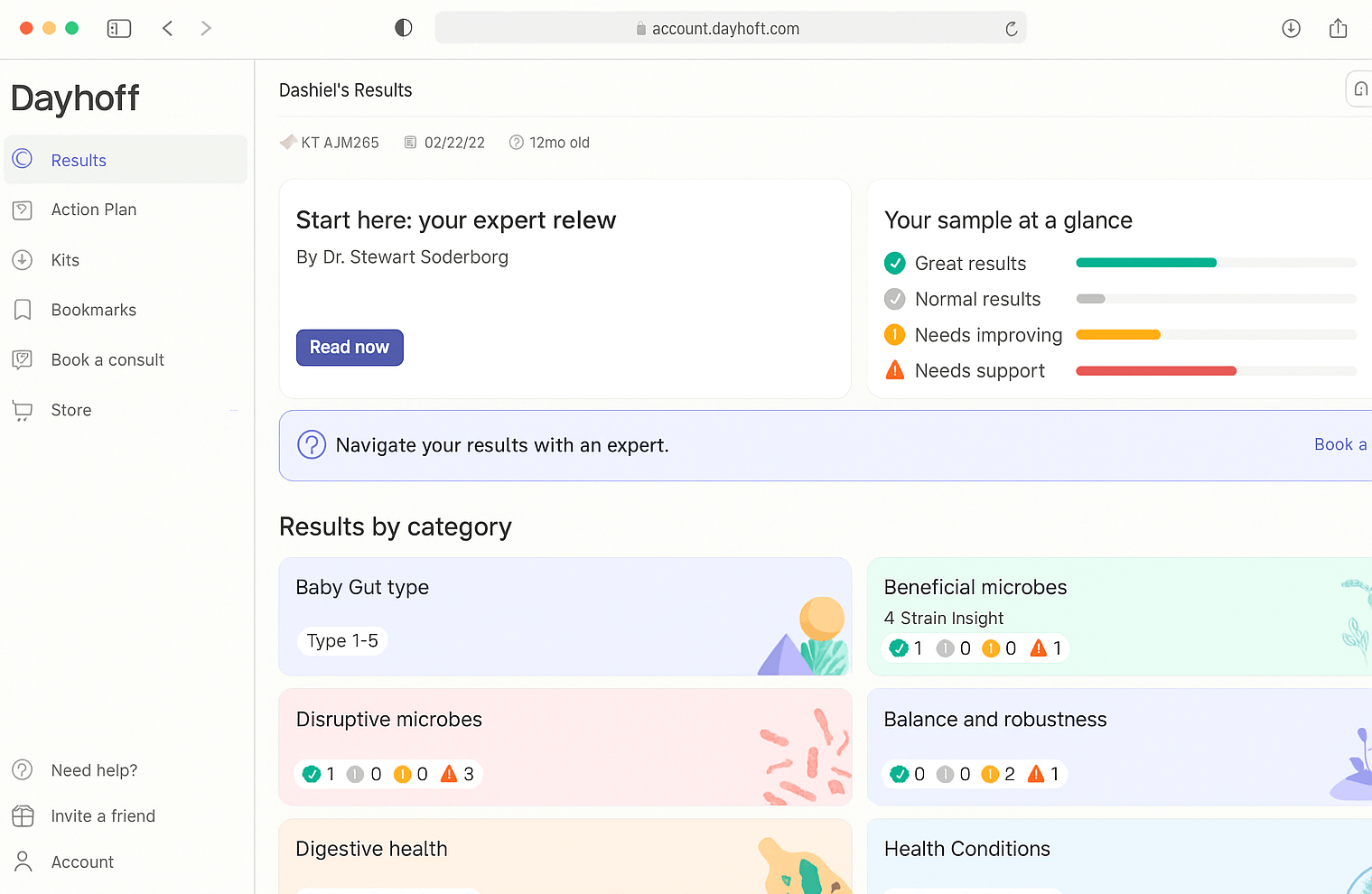
Discover the wide range of detailed microbiome categories and insights included in a Dayhoff report. For the best experience, view this list on a desktop.
Full List of Dayhoff Metrics:

Metagenomics is a field of genetics that focuses on the study of genetic material recovered directly from environmental samples. Unlike traditional microbiology, which often involves growing and identifying microorganisms in a lab, metagenomics bypasses the need for culturing organisms and instead analyzes the genetic material present in a sample. This allows for a broader, more comprehensive understanding of microbial diversity, as many microorganisms (such as bacteria, fungi, and viruses) cannot be easily cultured in a lab setting. Metagenomics is widely used in microbiome studies to understand the complex communities of microbes living in the human body, soil, water, and other environments.
Shotgun sequencing is a method used in genomics to sequence DNA. In this approach, the DNA from an organism or environmental sample is fragmented into numerous small pieces. These fragments are then sequenced, and the sequences are later assembled into longer sequences, allowing researchers to identify and analyze the entire genome. The "shotgun" analogy refers to the fact that the process randomly samples the DNA, similar to how shotgun pellets scatter in different directions. Shotgun sequencing is particularly useful in metagenomics because it can sequence a wide variety of organisms in a sample, even those that are difficult to culture.
Dayhoff, a company focused on microbiome testing, likely uses a form of metagenomic shotgun sequencing for microbiome analysis. The process typically involves:
Sample Collection: The user provides a stool, vaginal, or other biological sample using a swab or collection kit.
DNA Extraction: The DNA is extracted from the microbes present in the sample, which could include bacteria, fungi, viruses, and other microorganisms.
Sequencing: Shotgun sequencing or a similar sequencing method is applied to the DNA to break it into smaller fragments and identify the microbial DNA sequences.
Bioinformatics Analysis: The sequences are analyzed using computational tools to identify the various microbes in the sample and their relative abundances.
Reporting: Dayhoff then provides the user with a detailed report, highlighting the microbial species and their potential impact on health, such as influencing digestion, immunity, and overall well-being.
Dayhoff's swab or collection kits are specially designed to maintain the integrity of biological samples until they can be processed in the lab. To protect the DNA from degradation due to factors like heat, oxygen, or time, these kits incorporate stabilizing agents or preservatives. For instance, when collecting a stool sample, it is often combined with a buffer solution that helps preserve the DNA while preventing the growth of unwanted microbes. This ensures that the microbial community within the sample remains intact and accurately reflects the true composition for sequencing analysis.
Yes, microbiome tests, including those provided by Dayhoff, can identify a wide range of microbes, including parasites, worms, and other pathogens. Parasites like Giardia or Entamoeba histolytica and worms like Ascaris or Strongyloides can be detected using metagenomic sequencing. These pathogens might be identified based on their unique genetic markers, and their presence in the sample can help provide insights into gastrointestinal health, infections, and other conditions.
Yes, testing for Candida and Helicobacter pylori (H. pylori) is typically included in microbiome assessments.
Both of these microbes are common in the human body:
Candida: A type of yeast that can overgrow in certain conditions, causing infections like candidiasis, which can affect the digestive system, skin, or genitals.
H. pylori: A bacterium linked to stomach ulcers, gastritis, and even gastric cancer. It is commonly found in the stomach and can be detected using molecular techniques like PCR (Polymerase Chain Reaction) or sequencing.
These microbes are significant because their overgrowth or imbalance can lead to various health issues, and identifying their presence is essential for effective treatment.
Identifying microbes at the strain level is important because different strains of the same microbial species can have very different characteristics and effects on human health. For example:
Probiotic strains may offer health benefits, like improving gut health or boosting immunity.
Pathogenic strains of the same species could cause infections or other health problems.
Strain-level identification helps provide more accurate insights into how specific microbes might be influencing a person's health and can lead to more personalized and targeted treatments.
Both DNA and RNA sequencing have their strengths, and the choice between the two depends on the research question or the specific application:
DNA Sequencing: DNA sequencing identifies the genetic material present in the sample, providing a broad view of the microbial community's composition. It's useful for determining which organisms are present, their abundance, and overall diversity.
RNA Sequencing: RNA sequencing focuses on identifying which genes are actively being expressed. It can give insights into the functionality of the microbiome, showing which microbes are active and potentially involved in specific processes or disease mechanisms at the time of sampling.
In microbiome studies, DNA sequencing is more commonly used because it provides a comprehensive snapshot of the microbial community. However, RNA sequencing is helpful for understanding which microbes are actively influencing the body’s health.
The type of microbiome analysis that’s right for you depends on what you’re hoping to learn:
Gut Microbiome: If you’re interested in digestive health, food sensitivities, or conditions like IBS, Crohn’s disease, or leaky gut, a stool sample analysis might be best. This will give you insights into the composition and balance of your gut microbiome.
Vaginal Microbiome: For women experiencing recurrent yeast infections, bacterial vaginosis, or other vaginal health concerns, a vaginal swab might be more appropriate. This analysis can reveal microbial imbalances that affect vaginal health.
Comprehensive Health: If you're interested in understanding overall health and how your microbiome affects different aspects of your well-being, a more comprehensive test that covers both the gut and other body sites (like the skin or oral microbiome) might be ideal.
CLIA (Clinical Laboratory Improvement Amendments) and CAP (College of American Pathologists) certifications indicate that a laboratory meets high standards for quality and accuracy in testing.
CLIA Certification: CLIA is a set of regulations in the U.S. that ensures labs meet quality standards for testing. A CLIA-certified lab follows strict guidelines to ensure the accuracy, reliability, and timeliness of test results.
CAP Certification: CAP is an organization that sets accreditation standards for laboratories. CAP-certified labs undergo rigorous inspections to ensure that their testing processes and results meet the highest medical and scientific standards.
Both certifications ensure that the laboratory is qualified to perform diagnostic tests and that the results can be trusted for medical or health-related decisions.